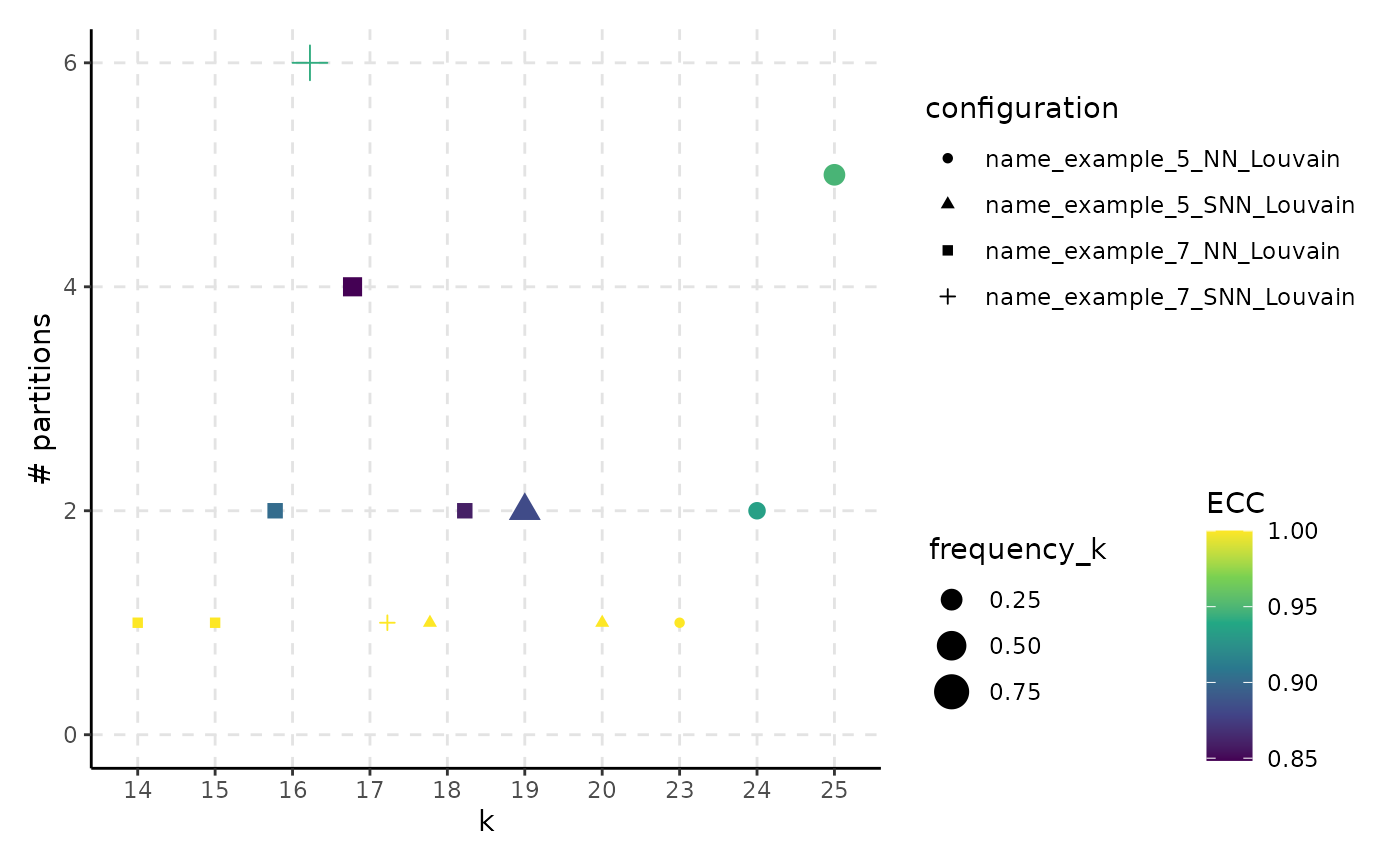
Relationship Between the Number of Clusters and the Number of Unique Partitions
Source:R/stability-3-graph-clustering.R
plot_k_n_partitions.RdFor each configuration provided in clust_object, display how many different partitions with the same number of clusters can be obtained by changing the seed.
Arguments
- clust_object
An object returned by the
assess_clustering_stabilitymethod.- colour_information
String that specifies the information type that will be illustrated using gradient colour: either
freq_partfor the frequency of the most common partition oreccfor the Element-Centric Consistency of the partitions obtained when the the number of clusters is fixed. Defaults toecc.- dodge_width
Used for adjusting the distance between the boxplots representing a clustering method. Defaults to
0.3.- pt_size_range
Indicates the minimum and the maximum size a point on the plot can have. Defaults to
c(1.5, 4).- summary_function
The function that will be used to summarize the distribution of the ECC values obtained for each number of clusters. Defaults to
median.- y_step
The step used for the y-axis. Defaults to
5.
Value
A ggplot2 object. The color gradient suggests the frequency of the most common partition relative to the total number of appearances of that specific number of clusters or the Element-Centric Consistency of the partitions. The size illustrates the frequency of the partitions with k clusters relative to the total number of partitions. The shape of the points indicates the clustering method.
Examples
set.seed(2024)
# create an artificial PCA embedding
pca_embedding <- matrix(runif(100 * 30), nrow = 100)
rownames(pca_embedding) <- paste0("cell_", seq_len(nrow(pca_embedding)))
colnames(pca_embedding) <- paste0("PC_", 1:30)
adj_matrix <- getNNmatrix(
RANN::nn2(pca_embedding, k = 10)$nn.idx,
10,
0,
-1
)$nn
rownames(adj_matrix) <- paste0("cell_", seq_len(nrow(adj_matrix)))
colnames(adj_matrix) <- paste0("cell_", seq_len(ncol(adj_matrix)))
# alternatively, the adj_matrix can be calculated
# using the `Seurat::FindNeighbors` function.
clust_diff_obj <- assess_clustering_stability(
graph_adjacency_matrix = adj_matrix,
resolution = c(0.5, 1),
n_repetitions = 10,
clustering_algorithm = 1:2,
verbose = FALSE
)
plot_k_n_partitions(clust_diff_obj)