Calculates the per-cell overlap of previously calculated marker genes.
Usage
marker_overlap(
markers1,
markers2,
clustering1,
clustering2,
n = 25,
overlap_type = "jsi",
rank_by = "-p_val",
use_sign = TRUE
)Arguments
- markers1
The first data frame of marker genes, must contain columns called 'gene' and 'cluster'.
- markers2
The second data frame of marker genes, must contain columns called 'gene' and 'cluster'.
- clustering1
The first vector of cluster assignments.
- clustering2
The second vector of cluster assignments.
- n
The number of top n markers (ranked by rank_by) to use when calculating the overlap.
- overlap_type
The type of overlap to calculated: must be one of 'jsi' for Jaccard similarity index and 'intersect' for intersect size.
- rank_by
A character string giving the name of the column to rank marker genes by. Note the sign here: to rank by lowest p-value, preface the column name with a minus sign; to rank by highest value, where higher value indicates more discriminative genes (for example power in the ROC test), no sign is needed.
- use_sign
A logical: should the sign of markers match for overlap calculations? So a gene must be a positive or a negative marker in both clusters being compared. If TRUE, markers1 and markers2 must have a 'avg_logFC' or 'avg_log2FC' column, from which the sign of the DE will be extracted.
Examples
suppressWarnings({
set.seed(1234)
library(Seurat)
data("pbmc_small")
# cluster with Louvain algorithm
pbmc_small <- FindClusters(pbmc_small, resolution = 0.8, verbose = FALSE)
# cluster with k-means
pbmc.pca <- Embeddings(pbmc_small, "pca")
pbmc_small@meta.data$kmeans_clusters <- kmeans(pbmc.pca, centers = 3)$cluster
# compare the markers
Idents(pbmc_small) <- pbmc_small@meta.data$seurat_clusters
louvain.markers <- FindAllMarkers(pbmc_small,
logfc.threshold = 1,
test.use = "t",
verbose = FALSE
)
Idents(pbmc_small) <- pbmc_small@meta.data$kmeans_clusters
kmeans.markers <- FindAllMarkers(pbmc_small,
logfc.threshold = 1,
test.use = "t",
verbose = FALSE
)
pbmc_small@meta.data$jsi <- marker_overlap(
louvain.markers, kmeans.markers,
pbmc_small@meta.data$seurat_clusters, pbmc_small@meta.data$kmeans_clusters
)
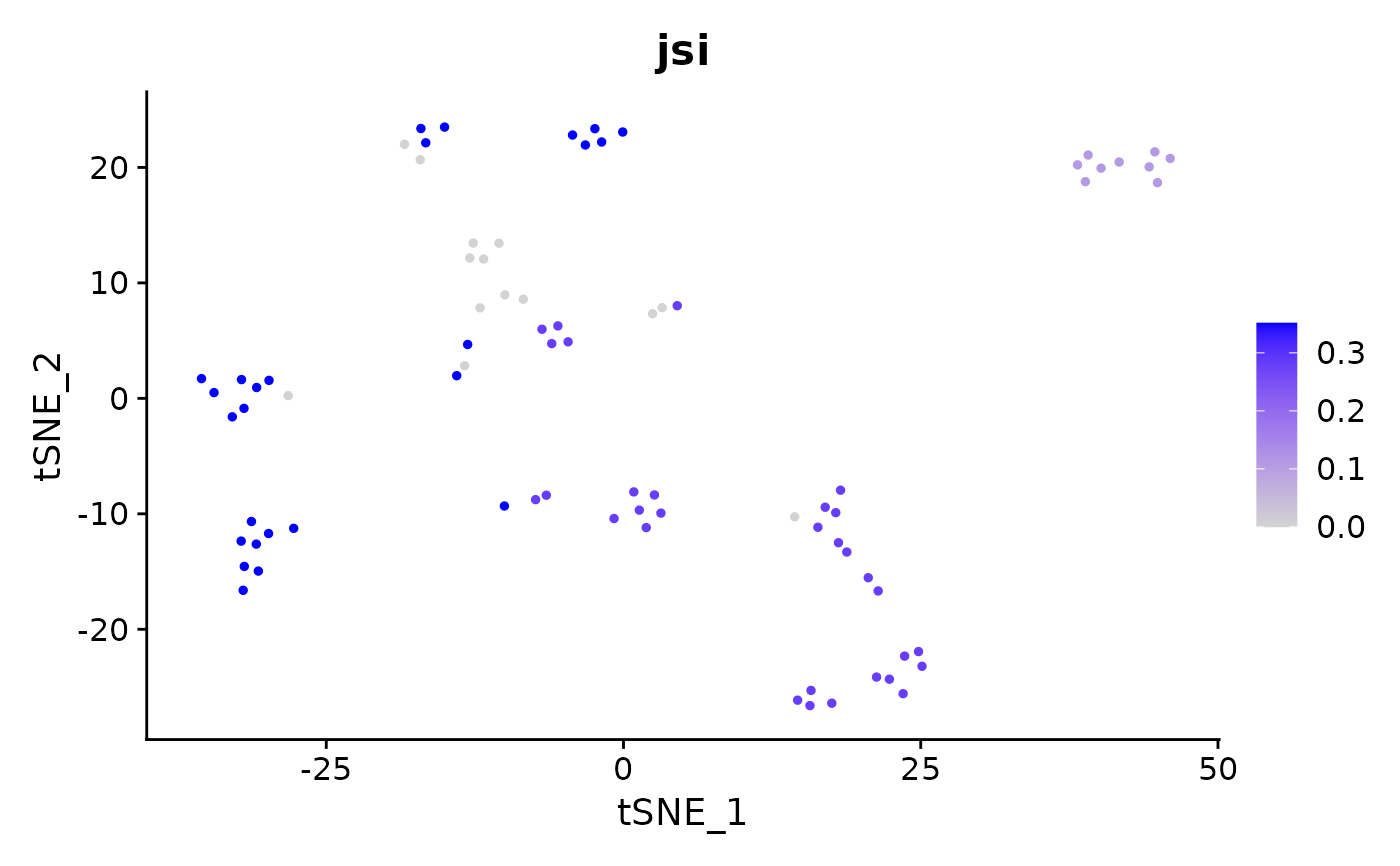
# which cells have the same markers, regardless of clustering?
FeaturePlot(pbmc_small, "jsi")
})
#> Loading required package: SeuratObject
#> Loading required package: sp
#>
#> Attaching package: ‘SeuratObject’
#> The following objects are masked from ‘package:base’:
#>
#> intersect, t